A. Make Surface ESP by G09
The total potential energy of the system for the induced dipole force field can be written as:
E = Ebnd + Enonbnd = Ebnd + Evdw + Ees + Eplz
The Ebnd term consists of bond, angle, dihedral angle terms, etc. The Enonbnd term consists of van der Waals, electrostatic, and polarization terms. The Eplz term is non-additive. The Enonbnd terms are focused on because they are directly involved in the interaction energy. The Lennard-Jones parameters of Evdw were essentially taken from the AMBER force field. These were partially adjusted by the introduction of the polarization term. The atomic charge parameters of Ees term are derived from the electrostatic potential(ESP) fitting. The ESP fitting of Gaussian 09 (G09) program 1 is used and the calculated ESP data are saved in the file. The saved data are used for the re-optimization of the charges using ESPopt program if nessessary. As shown in below, G09 is modified a little for the ESP data save. The atomic polarizability parameters of Eplz term are derived from the polarized one-electron potential (POP) fitting. The POPs are the difference between ESPs with and without an external test charge. The saved ESP data are also used for the POP fitting using POPopt program. Both fittings are important for building polarizable molecular block model.
Here, the modefication of G09 program is discrived in detail. Then the sample job for the modified G09 is shown. The data on the molecular surface point selection are also shown.
1. Modification of G09 to make restart ESP data
The subroutine SurGS3 and WriESP parts, which calculate the surface electrostatic potential (ESP) coordinates of a molecule and write it to an external file, were first implemented in the GSCF3 program.2 These routines can also be implemented in the Gaussian program series. Here, the implementation to G09 is explained.
The subroutines SurGS3 and WriESP must be implemented in G09 to use the ESPopt and POPopt programs to generate atomic charges and atomic polarizabilities, respectively.
When using the information on this website, please cite the four papers 2, 3, 4 and 5 shown below.
Three component programs, l601.F, l602.F, and utilam.F of G09, must be modified to incorporate the subroutines SurGS3 and WriESP. The program used was G09RevC.01. The modified parts are specifically displayed within the iframe.
l601.F
IOp(20) on Overlay 6 controles the electrostatic-potential drived charges. The option "1" is Merz-Kollman point selection. "2" is CHelp point selection. "3" is CHelpG point selection. Kosugi point selections are newly added. The option "6", "7", "8", and "9" represent 2.0, 1.7, 1.8, and 1.9 times the van der Waals radius, respectively.
Download
l602.F
Subroutine SurGS3 and WriESP are added in l602.F. Electrostatic potentials by Kosugi point selection are written in the restart file.
Download
utilam.F
VDW radius are defined in Subroutine GetVDW. Merz and Kollman values are used in Kosugi point selection.
Download
E = Ebnd + Enonbnd = Ebnd + Evdw + Ees + Eplz
The Ebnd term consists of bond, angle, dihedral angle terms, etc. The Enonbnd term consists of van der Waals, electrostatic, and polarization terms. The Eplz term is non-additive. The Enonbnd terms are focused on because they are directly involved in the interaction energy. The Lennard-Jones parameters of Evdw were essentially taken from the AMBER force field. These were partially adjusted by the introduction of the polarization term. The atomic charge parameters of Ees term are derived from the electrostatic potential(ESP) fitting. The ESP fitting of Gaussian 09 (G09) program 1 is used and the calculated ESP data are saved in the file. The saved data are used for the re-optimization of the charges using ESPopt program if nessessary. As shown in below, G09 is modified a little for the ESP data save. The atomic polarizability parameters of Eplz term are derived from the polarized one-electron potential (POP) fitting. The POPs are the difference between ESPs with and without an external test charge. The saved ESP data are also used for the POP fitting using POPopt program. Both fittings are important for building polarizable molecular block model.
Here, the modefication of G09 program is discrived in detail. Then the sample job for the modified G09 is shown. The data on the molecular surface point selection are also shown.
- Modification of G09 to make restart ESP data
- Sample input and output for modified G09
- Comparison with other point selections
- Kosugi point selection
- G09 program compile memo
1. Modification of G09 to make restart ESP data
The subroutine SurGS3 and WriESP parts, which calculate the surface electrostatic potential (ESP) coordinates of a molecule and write it to an external file, were first implemented in the GSCF3 program.2 These routines can also be implemented in the Gaussian program series. Here, the implementation to G09 is explained.
The subroutines SurGS3 and WriESP must be implemented in G09 to use the ESPopt and POPopt programs to generate atomic charges and atomic polarizabilities, respectively.
When using the information on this website, please cite the four papers 2, 3, 4 and 5 shown below.
Three component programs, l601.F, l602.F, and utilam.F of G09, must be modified to incorporate the subroutines SurGS3 and WriESP. The program used was G09RevC.01. The modified parts are specifically displayed within the iframe.
l601.F
IOp(20) on Overlay 6 controles the electrostatic-potential drived charges. The option "1" is Merz-Kollman point selection. "2" is CHelp point selection. "3" is CHelpG point selection. Kosugi point selections are newly added. The option "6", "7", "8", and "9" represent 2.0, 1.7, 1.8, and 1.9 times the van der Waals radius, respectively.
- Insert 4 lines of comment between lines 206 and 207 in Subroutine MulDrv.
- Insert 4 lines between lines 2500 and 2501 in Subroutine DQ.
- Insert 14 lines between lines 2583 and 2584 in Subroutine DQ.
Download
l602.F
Subroutine SurGS3 and WriESP are added in l602.F. Electrostatic potentials by Kosugi point selection are written in the restart file.
- Insert 6 lines of comment between lines 79 and 80 in Subroutine Prop1E.
- Insert "CelVec(3,3)," at line 95 in Subroutine Prop1E.
- Insert 6 lines between lines 625 and 626 in Subroutine Prop1E.
- Insert 22 lines between lines 1034 and 1035 in Subroutine Prop1E.
- Insert 3 lines of comment between lines 1275 and 1276 in Subroutine ESPFit.
- Add Subroutine SurGS3 to the bottom line of l602.F.
- Add Subroutine WriESP to the bottom line of Subroutine SurGS3.
Download
utilam.F
VDW radius are defined in Subroutine GetVDW. Merz and Kollman values are used in Kosugi point selection.
- Insert one lin of comment between lines 162008 and 162009 in Subroutine GetVDW.
- Insert 5 lines between lines 162042 and 162043 in Subroutine GetVDW.
Download
2. Sample input and output for modified G09
"opt_T_C_aT.gjf" is a sample input file for G09. The molecule to calculate is cytosine. After geometry optimization the ESP fitting is performed. IOp(20) on Overlay 6 controls the electrostatic-potential drived charges. To select the Kosugi point selections, the key word "pop=MK iop(6/20=8)" is required. The options "6", "7", "8", and "9" represent 2.0, 1.7, 1.8, and 1.9 times the van der Waals radius, respectively. The Kosugi point selections use the Merz-Kollman (MK) VDW radius.
Download
"runG09_opt.cm" is a sample C-shell script to run modified G09. The executable G09 is located at /usr/local/G09PN/g09/bsd/g09.login. Assume that the ".cshrc" file in home directory has the following line added:
"alias G09PN 'setenv g09root /usr/local/G09PN; source $g09root/g09/bsd/g09.login;time g09' "
The calculated surface potentials are written to the restart file "Rest_scf". (For MP2, the restart file name is "Rest_mp2".) Now rename the rest file to "opt_scf_C_aT.rest" to save it. This shell script can be run as a background job by using the command line:
"> cshrc runG09_opt.cm >& runG09_opt.log & "
Download
"opt_scf_C_aT.rest" is the restart file for ESP fitting containing atomic xyz coordinates (au), optimized charges (e), surface xyz coordinates (au), and ESPs (au) are included.
Download
"opt_T_C_aT.gjf" is a sample input file for G09. The molecule to calculate is cytosine. After geometry optimization the ESP fitting is performed. IOp(20) on Overlay 6 controls the electrostatic-potential drived charges. To select the Kosugi point selections, the key word "pop=MK iop(6/20=8)" is required. The options "6", "7", "8", and "9" represent 2.0, 1.7, 1.8, and 1.9 times the van der Waals radius, respectively. The Kosugi point selections use the Merz-Kollman (MK) VDW radius.
Download
"runG09_opt.cm" is a sample C-shell script to run modified G09. The executable G09 is located at /usr/local/G09PN/g09/bsd/g09.login. Assume that the ".cshrc" file in home directory has the following line added:
"alias G09PN 'setenv g09root /usr/local/G09PN; source $g09root/g09/bsd/g09.login;time g09' "
The calculated surface potentials are written to the restart file "Rest_scf". (For MP2, the restart file name is "Rest_mp2".) Now rename the rest file to "opt_scf_C_aT.rest" to save it. This shell script can be run as a background job by using the command line:
"> cshrc runG09_opt.cm >& runG09_opt.log & "
Download
"opt_scf_C_aT.rest" is the restart file for ESP fitting containing atomic xyz coordinates (au), optimized charges (e), surface xyz coordinates (au), and ESPs (au) are included.
Download
3. Comparison with Other Point Selections
The G09 ESP fit results of cytosine are shown in Table A1 and A2. The calculation level is wB97XD/aug-cc-pVTZ. The result of Merz-Kollman (MK) point selection (defauldt) is quite good. The Connolly surface is used in the MK selection. The relative root mean square deviation (rrmsd) is 5%. The reproducibility of the dipole moment is also quite good. The charges obtained from the ESP fit are shown in Table A2. The molecular surface points of Kosugi, Merz-Kollman, and GePol are shown below. These are perspective views with red dots on the front and black dots on the back. The rrmsd of CHelp and CHelpG are slightly heigher. The rrmsd of GePol is 5%. The rrmsd of Kosugi point selections are 3-4%. The reproducibility of the dipole moment is also quite good even with a single layer. The Kosugi selection uses the MK VDW radius. We are using the Kosugi 1.8.
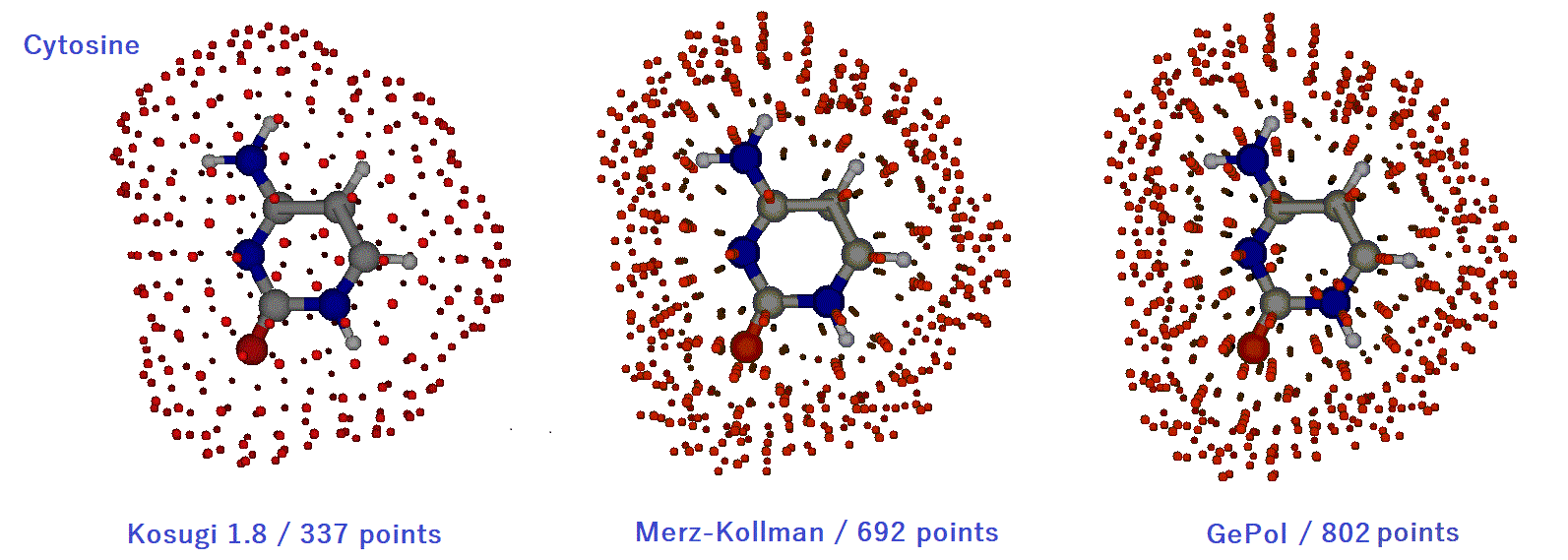
Molden is used to display molecules and molecular surface points.
The G09 ESP fit results of cytosine are shown in Table A1 and A2. The calculation level is wB97XD/aug-cc-pVTZ. The result of Merz-Kollman (MK) point selection (defauldt) is quite good. The Connolly surface is used in the MK selection. The relative root mean square deviation (rrmsd) is 5%. The reproducibility of the dipole moment is also quite good. The charges obtained from the ESP fit are shown in Table A2. The molecular surface points of Kosugi, Merz-Kollman, and GePol are shown below. These are perspective views with red dots on the front and black dots on the back. The rrmsd of CHelp and CHelpG are slightly heigher. The rrmsd of GePol is 5%. The rrmsd of Kosugi point selections are 3-4%. The reproducibility of the dipole moment is also quite good even with a single layer. The Kosugi selection uses the MK VDW radius. We are using the Kosugi 1.8.
| point selection | iop(6/20) | points | layer/grid | ESP fit RMSD (RRMSD) | μ x | μ y | μ z | μ Tot |
|---|---|---|---|---|---|---|---|---|
| wB97XD/ aug-cc-pVTZ | - | - | - | - | 4.84 | -4.57 | 0.00 | 6.65 |
| Merz-Kollman | 1 | 692 | 4 | 0.0015 (4.74 %) | 4.85 | -4.56 | 0.00 | 6.66 |
| CHelp | 2 | 849 | 5 | 0.0035 (15.93 %) | 4.83 | -4.54 | 0.00 | 6.63 |
| CHelpG | 3 | 9586 | grid | 0.0029 (9.62 %) | 4.87 | -4.55 | 0.00 | 6.67 |
| GePol | 4 | 802 | 4 | 0.0013 (4.62 %) | 4.87 | -4.57 | -0.00 | 6.68 |
| Kosugi 1.7 | 7 | 333 | 1 | 0.0012 (4.00 %) | 4.84 | -4.59 | -0.00 | 6.67 |
| Kosugi 1.8 | 8 | 337 | 1 | 0.0010 (3.56 %) | 4.85 | -4.58 | -0.00 | 6.67 |
| Kosugi 1.9 | 9 | 338 | 1 | 0.0008 (3.16 %) | 4.84 | -4.59 | -0.00 | 6.67 |
| Kosugi 2.0 | 6 | 345 | 1 | 0.0007 (2.86 %) | 4.84 | -4.59 | -0.00 | 6.67 |
| Atom | Merz-Kollman | CHelp | CHelpG | GePol | Kosugi 1.7 | Kosugi 1.8 | Kosugi 1.9 | Kosugi 2.0 |
|---|---|---|---|---|---|---|---|---|
| H1 | 0.358 | 0.312 | 0.341 | 0.352 | 0.368 | 0.369 | 0.371 | 0.367 |
| N1 | -0.645 | -0.496 | -0.615 | -0.648 | -0.699 | -0.707 | -0.718 | -0.731 |
| C6 | 0.283 | 0.168 | 0.280 | 0.316 | 0.353 | 0.361 | 0.379 | 0.393 |
| H6 | 0.131 | 0.107 | 0.156 | 0.124 | 0.119 | 0.120 | 0.118 | 0.117 |
| C5 | -0.768 | -0.529 | -0.648 | -0.827 | -0.862 | -0.883 | -0.907 | -0.924 |
| H5 | 0.241 | 0.173 | 0.189 | 0.256 | 0.259 | 0.263 | 0.268 | 0.272 |
| C4 | 1.070 | 0.961 | 1.023 | 1.123 | 1.189 | 1.218 | 1.239 | 1.252 |
| N4 | -1.072 | -1.083 | -1.045 | -1.099 | -1.149 | -1.168 | -1.175 | -1.178 |
| H41 | 0.451 | 0.445 | 0.437 | 0.458 | 0.472 | 0.478 | 0.479 | 0.479 |
| H42 | 0.443 | 0.438 | 0.429 | 0.449 | 0.461 | 0.466 | 0.468 | 0.469 |
| N3 | -0.845 | -0.772 | -0.833 | -0.871 | -0.902 | -0.912 | -0.921 | -0.929 |
| C2 | 0.990 | 0.932 | 0.990 | 1.019 | 1.038 | 1.040 | 1.043 | 1.051 |
| O2 | -0.639 | -0.655 | -0.653 | -0.650 | -0.647 | -0.645 | -0.643 | -0.644 |

Molden is used to display molecules and molecular surface points.
4. Kosugi point selection
To select the Kosugi point selections, the key word "pop=MK iop(6/20=8)" is required. The options "6", "7", "8", and "9" represent 2.0, 1.7, 1.8, and 1.9 times the van der Waals radius, respectively. The Kosugi point selections use the MK VDW radius. For example, the VDW radii of the H and C atoms are 1.2 Å and 1.5 Å, respectively.
When the VDW radius is multiplied by 1.8, 111 points are placed on the surface of H atom and 135 points are placed on the surface of C atom. As shown in the figure below, the larger the atomic surface, the greater the number of points to be arranged. These are perspective views with red dots on the front and black dots on the back.
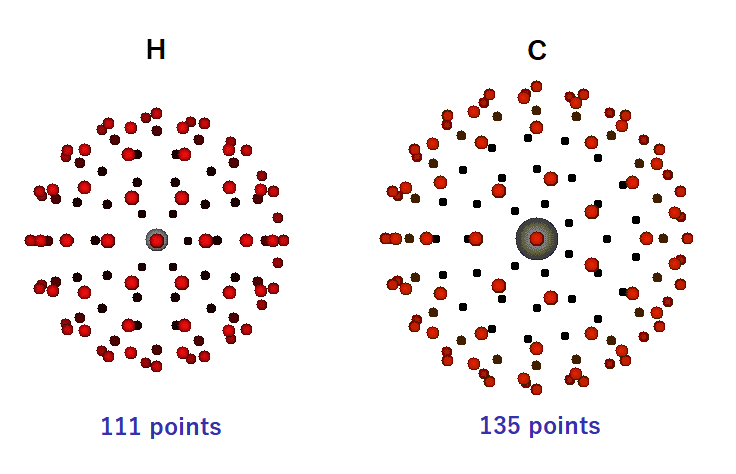
If we compare the H sphere to the Earth, there are 17 points on the equator and 1 point each on the north and south poles. The distance between the North Pole and the equator is divided into 5 equal parts of 18 degrees each, and 6, 10, 14, and 16 points are placed in each. The southern hemisphere is similarly scored. In this way, 111 points are placed on the H sphere as evenly as possible. Unfortunately, Kosugi point selection cannot arrange vertices evenly like the regular icosahedron, and the directionality is not even.
In the case of molecule, we remove points that overlap between atoms. The latest revision of subroutine SurGS3 shown above removes one of the too close points. It reduces the number of points and distributes the points more evenly. The cytosine surface is shown above. It is down 10 points from the original SurGS3.
To select the Kosugi point selections, the key word "pop=MK iop(6/20=8)" is required. The options "6", "7", "8", and "9" represent 2.0, 1.7, 1.8, and 1.9 times the van der Waals radius, respectively. The Kosugi point selections use the MK VDW radius. For example, the VDW radii of the H and C atoms are 1.2 Å and 1.5 Å, respectively.
When the VDW radius is multiplied by 1.8, 111 points are placed on the surface of H atom and 135 points are placed on the surface of C atom. As shown in the figure below, the larger the atomic surface, the greater the number of points to be arranged. These are perspective views with red dots on the front and black dots on the back.

If we compare the H sphere to the Earth, there are 17 points on the equator and 1 point each on the north and south poles. The distance between the North Pole and the equator is divided into 5 equal parts of 18 degrees each, and 6, 10, 14, and 16 points are placed in each. The southern hemisphere is similarly scored. In this way, 111 points are placed on the H sphere as evenly as possible. Unfortunately, Kosugi point selection cannot arrange vertices evenly like the regular icosahedron, and the directionality is not even.
In the case of molecule, we remove points that overlap between atoms. The latest revision of subroutine SurGS3 shown above removes one of the too close points. It reduces the number of points and distributes the points more evenly. The cytosine surface is shown above. It is down 10 points from the original SurGS3.
5. G09 program compile memo
Install the PGI compiler for compiling the G09 source code in advance. There is a recommended version of the PGI compiler for each revision of G09, so you will need to download the appropriate version of the PGI compiler. For G09 RevC.01, download version 11.8 (pgilinux-2011-118.tar.gz) from the PGI 2011 Release Archive (https://www.pgroup.com/).
Prepare to install the PGI compiler
Create the /scr/pgi directory in the scratch file, and expand the downloaded pgilinux-2011-118.tar.gz there
Install PGI in English environment
Get a new license (license.dat) from https://www.pgroup.com/account/login.php
Reset and start FlexNet license manager
Compile G09
Configure the environment for using G09 in your home directory
Install the PGI compiler for compiling the G09 source code in advance. There is a recommended version of the PGI compiler for each revision of G09, so you will need to download the appropriate version of the PGI compiler. For G09 RevC.01, download version 11.8 (pgilinux-2011-118.tar.gz) from the PGI 2011 Release Archive (https://www.pgroup.com/).
Prepare to install the PGI compiler
1) Stop the FlexNet license manager if necessary.
2) If there is an existing pgi compiler in the /opt/pgi directory, rename it and take a backup.
3) /opt/pgi is the default directory for the pgi compiler, so create an empty pgi directory.
# /etc/init.d/lmgrd-pgi stop
#cd /opt
#mv pgi pgi_old
#mkdir pgi
Set PGI environment variables in .cshrc for csh2) If there is an existing pgi compiler in the /opt/pgi directory, rename it and take a backup.
3) /opt/pgi is the default directory for the pgi compiler, so create an empty pgi directory.
# /etc/init.d/lmgrd-pgi stop
#cd /opt
#mv pgi pgi_old
#mkdir pgi
Add four lines in "/root/.cshrc" file
setenv PGI /opt/pgi
set path = ( $PGI/linux86-64/11.8/bin $path )
setenv MANPATH "$MANPATH":$PGI/linux86-64/11.8/man
setenv LM_LICENSE_FILE $PGI/license.dat
Flush root environment variables setenv PGI /opt/pgi
set path = ( $PGI/linux86-64/11.8/bin $path )
setenv MANPATH "$MANPATH":$PGI/linux86-64/11.8/man
setenv LM_LICENSE_FILE $PGI/license.dat
#source .cshrc
Create the /scr/pgi directory in the scratch file, and expand the downloaded pgilinux-2011-118.tar.gz there
#cd /scr
#ls
... pgilinux-2011-118.tar.gz ...
#mkdir pgi
#cd pgi
#tar zxvf ../pgilinux-2011-118.tar.gz
#ls
... pgilinux-2011-118.tar.gz ...
#mkdir pgi
#cd pgi
#tar zxvf ../pgilinux-2011-118.tar.gz
Install PGI in English environment
#setenv LANG C
#./install
License Agreement
Do you accept terms? [accept]
1 Select Single System Install [1]
Installation directory? [/opt/pgi]
Install the ACML? [y]
If you agree these terms? [acccept]
Install CUDA Toolkit Components? [y]
Do you accept these terms? [accept]
Instal JAVA JRE 6.0_21? [yes]
...
Do you wish to generate license keys or configure license service? [n]
Do you want the files in the install directory to be read-only? [n]
See /opt/pgi/ #./install
License Agreement
Do you accept terms? [accept]
1 Select Single System Install [1]
Installation directory? [/opt/pgi]
Install the ACML? [y]
If you agree these terms? [acccept]
Install CUDA Toolkit Components? [y]
Do you accept these terms? [accept]
Instal JAVA JRE 6.0_21? [yes]
...
Do you wish to generate license keys or configure license service? [n]
Do you want the files in the install directory to be read-only? [n]
#cd /opt/pgi
#ls
INSTALL.txt SUBSCRIPTION_SERVICE license.info linux86 linux86-64
#cat license.info
System information:
FLEXnet hostid "xxxxxxxxxfe xxxxxxxxxxff"
Hostname xxxxxx6b
Installation /opt/pgi
PGI Release 11.8
#ls
INSTALL.txt SUBSCRIPTION_SERVICE license.info linux86 linux86-64
#cat license.info
System information:
FLEXnet hostid "xxxxxxxxxfe xxxxxxxxxxff"
Hostname xxxxxx6b
Installation /opt/pgi
PGI Release 11.8
Get a new license (license.dat) from https://www.pgroup.com/account/login.php
Email address [xxxx@xxxx-u.ac.jp]
Password [........]
LOGIN
License Management
PIN xxxx33 PIN code xxxx-xxxx-xxxx-xxxx Accounts tied xxxx@xxxx-u.ac.jp
Generate Software License Keys for PIN xxxx33
To transfer your license keys to another host, first delete this key and then create a new key with the new hostid.
Delete license key
It will be available the next day, so get the license file with the new hostname.
Generate Software License Keys for PIN xxxx33
Update/Display license key
Generate Software License keys for PIN xxxx33
I accept ...
Hostname [xxxx6b]
FLEXnet hostsid [xxxxxxxxxxfe]
GENERATE LICENSE KEY
Generate Software License Keys for PIN xxxx33
Save keys as a file on my computer
SERVER xxxxxx6b xxxxxxxxxxfe xxxx0
DAEMON pgroupd
PACKAGE PGI2013-xxxx33 pgrouped ...
...
Place the license.dat in the /opt/pgi directory Password [........]
LOGIN
License Management
PIN xxxx33 PIN code xxxx-xxxx-xxxx-xxxx Accounts tied xxxx@xxxx-u.ac.jp
Generate Software License Keys for PIN xxxx33
To transfer your license keys to another host, first delete this key and then create a new key with the new hostid.
Delete license key
It will be available the next day, so get the license file with the new hostname.
Generate Software License Keys for PIN xxxx33
Update/Display license key
Generate Software License keys for PIN xxxx33
I accept ...
Hostname [xxxx6b]
FLEXnet hostsid [xxxxxxxxxxfe]
GENERATE LICENSE KEY
Generate Software License Keys for PIN xxxx33
Save keys as a file on my computer
SERVER xxxxxx6b xxxxxxxxxxfe xxxx0
DAEMON pgroupd
PACKAGE PGI2013-xxxx33 pgrouped ...
...
Reset and start FlexNet license manager
Check Hostname in /etc/hosts
#ifconfig
Check eth0 port IP address
Register the FlexNet startup script (lmgrd.rc) in the system #ifconfig
Check eth0 port IP address
#cp $pgi/linux86-64/11.8/bin/lmgrd.rc /etc/init.d/lmgrd-pgi
#chkconfig --list | grep lmgrd-pgi
lmgrd-pgi 0:off 1:off 2:on 3:on 4:on 5:on 6:off
Check 3 and 5 should be on
#/etc/init.d/lmgrd-pgi start
#chkconfig --list | grep lmgrd-pgi
lmgrd-pgi 0:off 1:off 2:on 3:on 4:on 5:on 6:off
Check 3 and 5 should be on
#/etc/init.d/lmgrd-pgi start
Compile G09
#cd /usr/local/G09P_new/g09
# bsd/bldg09 > & make_11.8.log &
It takes several hours.
# bsd/bldg09 > & make_11.8.log &
It takes several hours.
Configure the environment for using G09 in your home directory
Add following a line in .cshrc.
alias G09PN 'setenv g09root /usr/local/G09P_new; source $g09root/g09/bsd/g09.login;time g09'
> source .cshrc
alias G09PN 'setenv g09root /usr/local/G09P_new; source $g09root/g09/bsd/g09.login;time g09'
> source .cshrc
References
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson et al.
Gaussian 09, Revision C.01, Gaussian, Inc., Wallingford CT, 2010.
- S. Nakagawa and N. Kosugi,
Polarized one-electron potentials fitted by multicenter polarizabilities and hyperpolarizabilities. Ab initio SCF-CI calculation of water.
Chem. Phys. Letters, 210, 180-186 (1993).
- S. Nakagawa, Polarizable Model Potential Function for Nucleic Acid Bases. J. Comput. Chem., 1538-1550 (2007).
- S. Nakagawa, P. Mark, H. Agren,
Recipe of Polarized One-electron Potential Optimization for Development of Polarizable Force Fields.
J. Chem. Theory Comput., 28, 1947-1959 (2007).
- S. Nakagawa, A. Kimura, Y. Okamoto,
Polarizable Molecular Block Model: Toward the Development of an Induced Dipole Force Field for DNA.
J. Phys. Chem. B, 126, 10646-10661 (2022).